8 clusterProfiler包做GO,KEGG富集
8.1 安装clusterProfiler包及所需文件
# 安装clusterProfiler包。
# 可能会报错,可以下载到本地安装,可参考https://zhuanlan.zhihu.com/p/436671645
BiocManager::install("clusterProfiler")
# 安装所需的物种数据库注释文件
BiocManager::install("org.Hs.eg.db") # 人的注释数据库。其他物种的可在https://bioconductor.org/packages/3.6/data/annotation/找到8.2 ID Mapping
# 通过bitr函数,做Symbol,Gene ID,uniprot ID 等之间的转换
# 支持如下类型之间的相互转换:ACCNUM, ALIAS, ENSEMBL, ENSEMBLPROT, ENSEMBLTRANS, ENTREZID, ENZYME, EVIDENCE, EVIDENCEALL, GENENAME, GO, GOALL, IPI, MAP, OMIM, ONTOLOGY, ONTOLOGYALL, PATH, PFAM, PMID, PROSITE, REFSEQ, SYMBOL, UCSCKG, UNIGENE, UNIPROT
library(clusterProfiler)
symbolList <- c("GPX3", "GLRX", "LBP", "CRYAB", "DEFB1", "HCLS1", "SOD2", "HSPA2",
"ORM1", "IGFBP1", "PTHLH", "GPC3", "IGFBP3","TOB1", "MITF", "NDRG1",
"NR1H4", "FGFR3", "PVR", "IL6", "PTPRM", "ERBB2", "NID2", "LAMB1",
"COMP", "PLS3", "MCAM", "SPP1", "LAMC1", "COL4A2", "COL4A1", "MYOC",
"ANXA4", "TFPI2", "CST6", "SLPI", "TIMP2", "CPM", "GGT1", "NNMT",
"MAL", "EEF1A2", "HGD", "TCN2", "CDA", "PCCA", "CRYM", "PDXK",
"STC1", "WARS", "HMOX1", "FXYD2", "RBP4", "SLC6A12", "KDELR3", "ITM2B")
eg <- bitr(symbolList,
fromType="SYMBOL", # 定义输入类型
toType=c("ENTREZID","ENSEMBL","UNIPROT"), # 输出何种类型
OrgDb="org.Hs.eg.db") # (人)物种数据库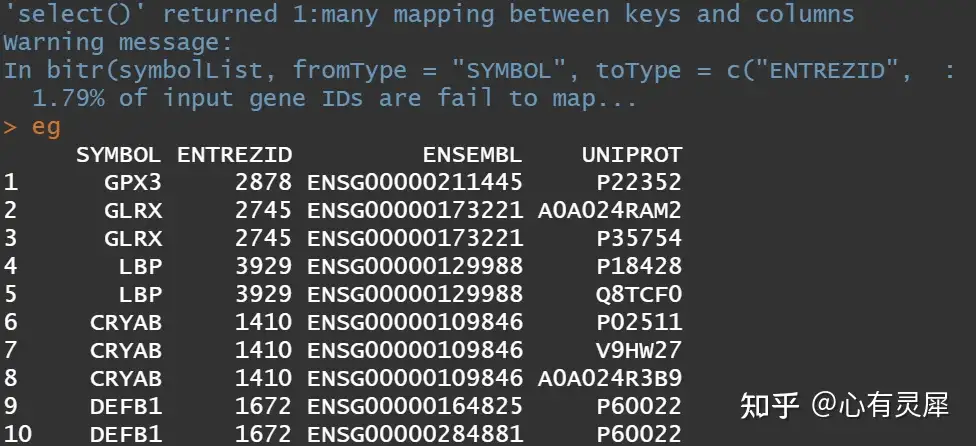
8.3 GO富集
genelist <- eg$ENTREZID
genelist = genelist[-duplicated(genelist)] # 去重
go <- enrichGO(genelist,
OrgDb = org.Hs.eg.db, # 选择对应物种的数据库
ont='ALL', # 选择GO富集的种类"BP", "MF", "CC" OR "ALL"
pAdjustMethod = 'BH', # FDR算法
pvalueCutoff = 1, # 过滤p值小于多少的条目,1为不过滤
qvalueCutoff = 1, # 过滤q值小于多少的条目,1为不过滤
keyType = 'ENTREZID', # 定义输入类型为NCBI的Gene ID
readable = T # 是否将结果表格中的富集条目改为Gene Symbol,增加可读性
)
head(go)
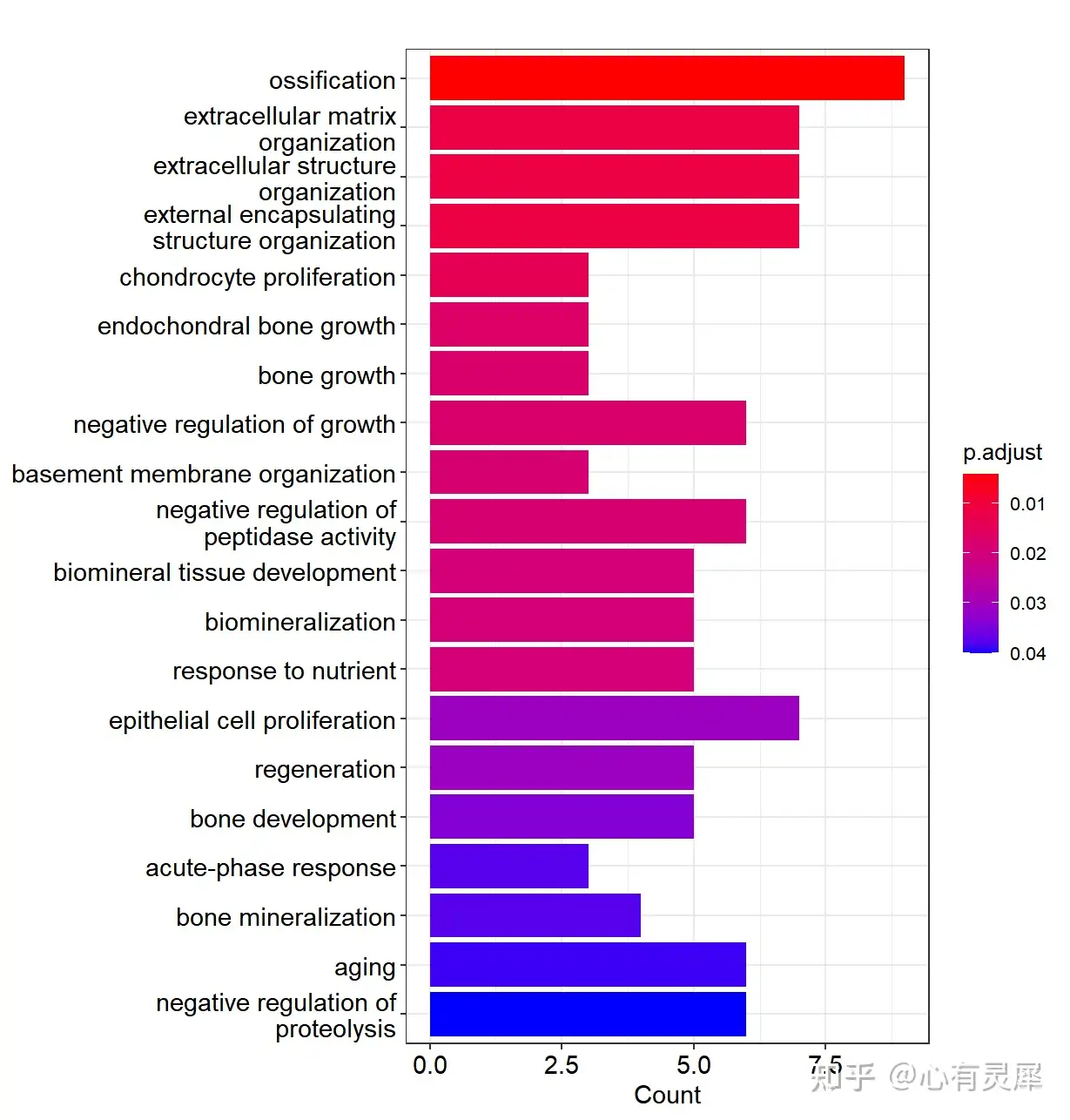
barplot(go,showCategory=20,drop=T) # 绘制富集条形图
dotplot(go,showCategory=20) # 绘制富集气泡图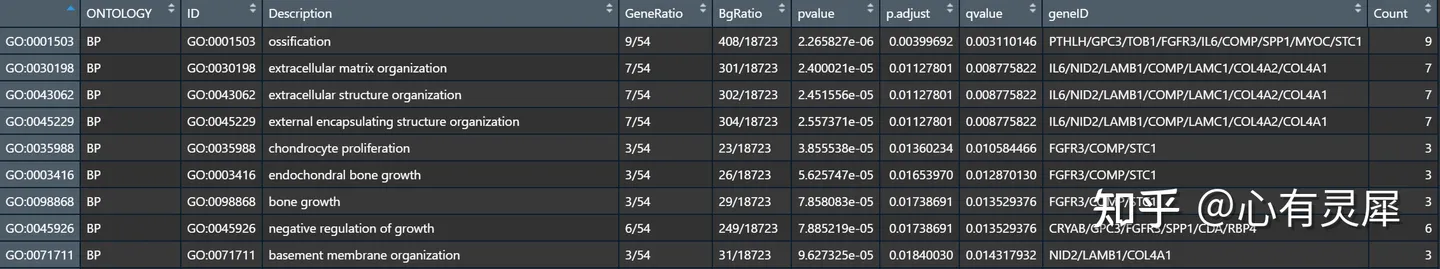

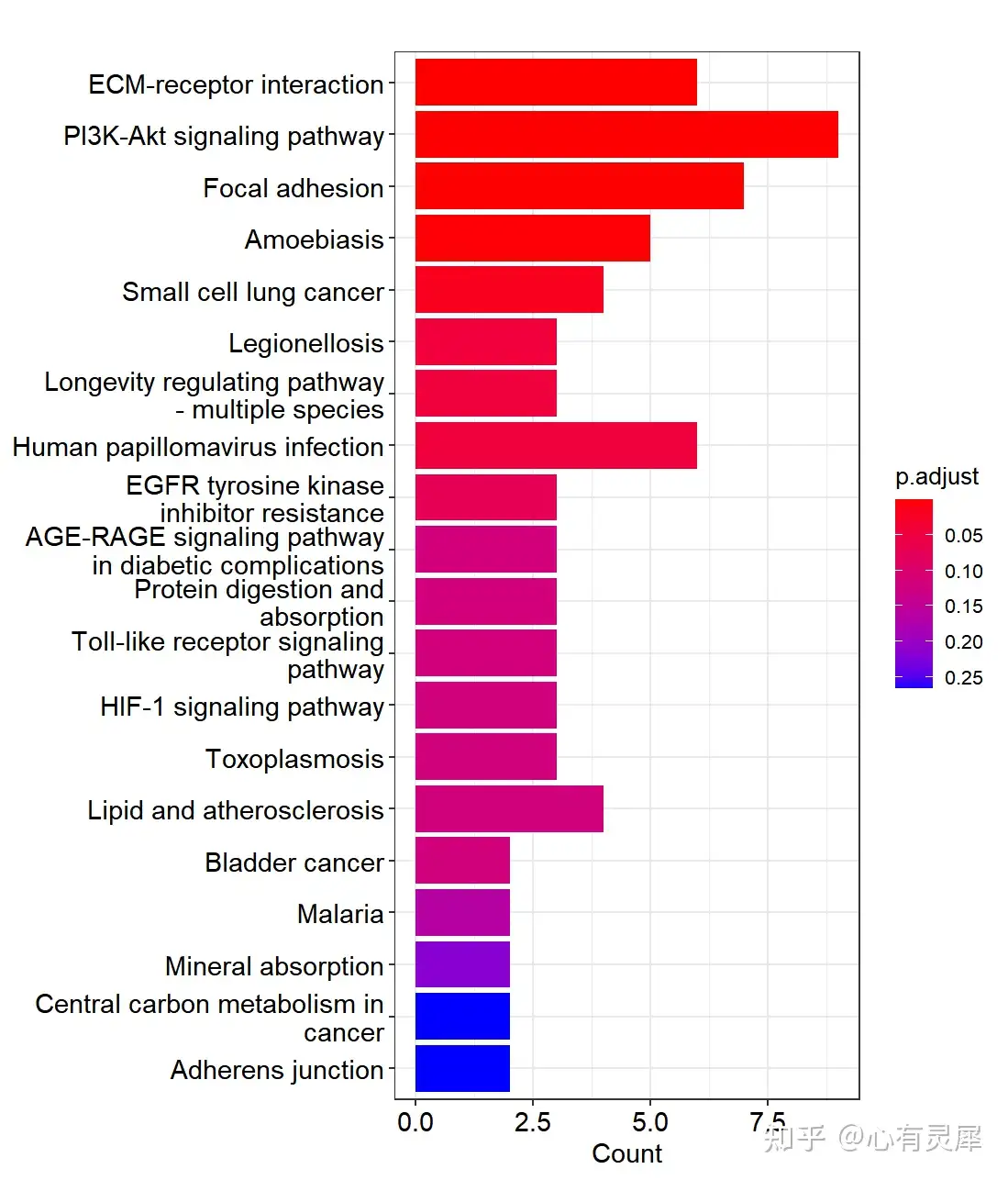
8.4 KEGG富集
kegg <- enrichKEGG(genelist,
organism = 'hsa', # 选择对应物种的数据库,通过如下网址,获取对应物种的三字母缩写http://www.genome.jp/kegg/catalog/org_list.html
keyType = 'kegg', # 定义输入类型
pvalueCutoff = 1, # 过滤p值小于多少的条目,1为不过滤
qvalueCutoff = 1, # 过滤q值小于多少的条目,1为不过滤
pAdjustMethod = 'BH', # FDR算法
use_internal_data = F # 是否使用KEGG.db本地数据库
)
head(kegg)
barplot(kegg,showCategory=20,drop=T) # 绘制富集条形图
dotplot(kegg,showCategory=20) # 绘制富集气泡图