14 三维PCA(可交互)
基本过程:
1,通过R语言自带的函数prcomp(),计算绘制PCA点图的坐标位置
2,通过plotly包绘制三维散点图
结果预览如下
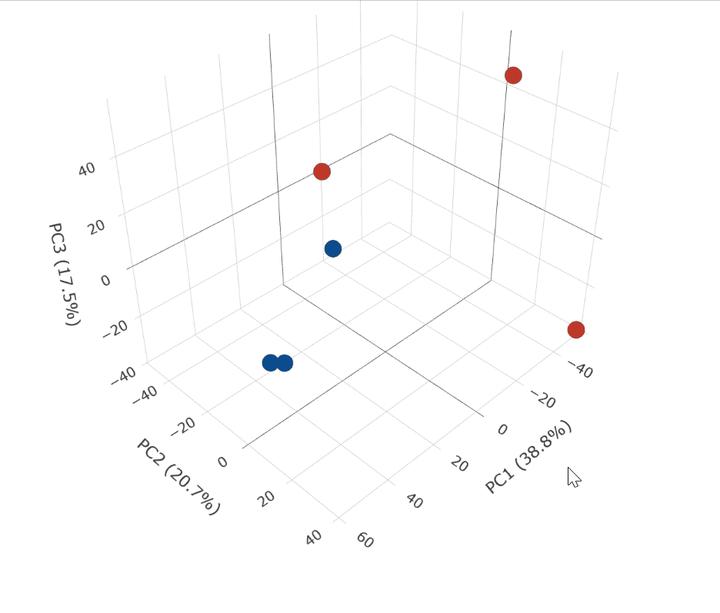
代码如下:
# 加载R包,没有安装请先安装 install.packages("包名")
library(plotly)
library(tidyverse)
library(htmlwidgets)
# 读取PCA数据文件
df = read.delim("https://www.bioladder.cn/shiny/zyp/bioladder2/demoData/PCA3_HTML/data.txt",# 这里读取了网络上的demo数据,将此处换成你自己电脑里的文件
header = T, # 指定第一行是列名
row.names = 1 # 指定第一列是行名
)
df=t(df) # 对数据进行转置,如果想对基因分组则不用转置
# 读取样本分组数据文件
dfGroup = read.delim("https://www.bioladder.cn/shiny/zyp/bioladder2/demoData/PCA3_HTML/class.txt",
header = T,
row.names = 1
)
# PCA计算
pca_result <- prcomp(df,
scale=T # 一个逻辑值,指示在进行分析之前是否应该将变量缩放到具有单位方差
)
pca_result$x<-data.frame(pca_result$x)
#计算PC值,并替换列名,用来替换坐标轴上的标签
pVar <- pca_result$sdev^2/sum(pca_result$sdev^2)
pVar = round(pVar,digits = 3)
colnames(pca_result$x)[1] = paste0("PC1 (",as.character(pVar[1] * 100 ),"%)")
colnames(pca_result$x)[2] = paste0("PC2 (",as.character(pVar[2] * 100 ),"%)")
colnames(pca_result$x)[3] = paste0("PC3 (",as.character(pVar[3] * 100 ),"%)")
pca_result$x = pca_result$x %>%
rownames_to_column()
# 绘图
fig <- plot_ly(pca_result$x,
text = ~paste0(rowname),
x = ~pca_result$x[,2],
y = ~pca_result$x[,3],
z = ~pca_result$x[,4],
color = ~dfGroup[,1],
colors = c('#BF382A', '#0C4B8E')
)
fig <- fig %>% add_markers()
fig <- fig %>% layout(scene = list(xaxis = list(title = colnames(pca_result$x)[2]),
yaxis = list(title = colnames(pca_result$x)[3]),
zaxis = list(title = colnames(pca_result$x)[4])))
fig <- fig %>% config(displaylogo = FALSE)
fig
# 保存为网页
htmlwidgets::saveWidget(fig,"1.html")